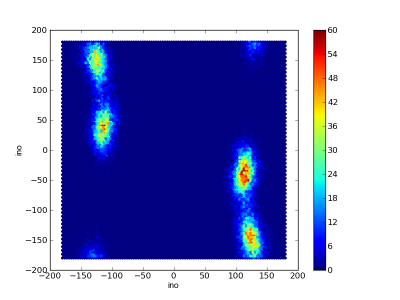
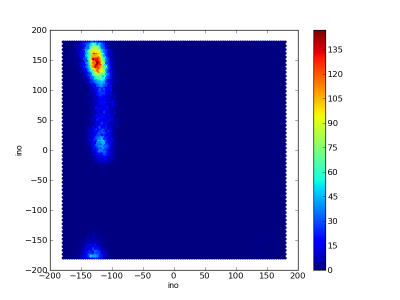
Dihedral debugging
Glycine tripeptide
Crude 50 ns simulation of Gly(3) in vacuum.
Ramachandran
Files
mdp file:
; ; File 'gly3_fullmd__1287526816.mdp' was generated ; By user: ms872 (1005) ; On host: nursie ; At date: Tue Oct 19 23:20:28 2010 ; ; VARIOUS PREPROCESSING OPTIONS ; Preprocessor information: use cpp syntax. ; e.g.: -I/home/joe/doe -I/home/mary/hoe include = -I../top ; e.g.: -DI_Want_Cookies -DMe_Too define = ; RUN CONTROL PARAMETERS integrator = sd ; Start time and timestep in ps tinit = 0 dt = 0.002 nsteps = 25000000 ; For exact run continuation or redoing part of a run ; Part index is updated automatically on checkpointing (keeps files separate) simulation_part = 1 init_step = 0 ; mode for center of mass motion removal comm-mode = Linear ; number of steps for center of mass motion removal nstcomm = 1 ; group(s) for center of mass motion removal comm-grps = ; LANGEVIN DYNAMICS OPTIONS ; Friction coefficient (amu/ps) and random seed bd-fric = 0 ld-seed = 1993 ; ENERGY MINIMIZATION OPTIONS ; Force tolerance and initial step-size emtol = 10 emstep = 0.01 ; Max number of iterations in relax_shells niter = 20 ; Step size (ps^2) for minimization of flexible constraints fcstep = 0 ; Frequency of steepest descents steps when doing CG nstcgsteep = 1000 nbfgscorr = 10 ; TEST PARTICLE INSERTION OPTIONS rtpi = 0.05 ; OUTPUT CONTROL OPTIONS ; Output frequency for coords (x), velocities (v) and forces (f) nstxout = 1000 nstvout = 1000 nstfout = 0 ; Output frequency for energies to log file and energy file nstlog = 1000 nstenergy = 1000 ; Output frequency and precision for xtc file nstxtcout = 1000 xtc-precision = 1000 ; This selects the subset of atoms for the xtc file. You can ; select multiple groups. By default all atoms will be written. xtc_grps = protein ; Selection of energy groups energygrps = ; NEIGHBORSEARCHING PARAMETERS ; nblist update frequency nstlist = 10 ; ns algorithm (simple or grid) ns_type = grid ; Periodic boundary conditions: xyz, no, xy pbc = xyz periodic_molecules = no ; nblist cut-off rlist = 0.8 ; OPTIONS FOR ELECTROSTATICS AND VDW ; Method for doing electrostatics coulombtype = cut-off rcoulomb-switch = 0 rcoulomb = 1.4 ; Relative dielectric constant for the medium and the reaction field epsilon_r = 1 epsilon_rf = 1 ; Method for doing Van der Waals vdwtype = cut-off ; cut-off lengths rvdw-switch = 0 rvdw = 1.4 ; Apply long range dispersion corrections for Energy and Pressure DispCorr = No ; Extension of the potential lookup tables beyond the cut-off table-extension = 1 ; Seperate tables between energy group pairs energygrp_table = ; Spacing for the PME/PPPM FFT grid fourierspacing = 0.12 ; FFT grid size, when a value is 0 fourierspacing will be used fourier_nx = 0 fourier_ny = 0 fourier_nz = 0 ; EWALD/PME/PPPM parameters pme_order = 4 ewald_rtol = 1e-05 ewald_geometry = 3d epsilon_surface = 0 optimize_fft = no ; IMPLICIT SOLVENT ALGORITHM implicit_solvent = No ; GENERALIZED BORN ELECTROSTATICS ; Algorithm for calculating Born radii gb_algorithm = Still ; Frequency of calculating the Born radii inside rlist nstgbradii = 1 ; Cutoff for Born radii calculation; the contribution from atoms ; between rlist and rgbradii is updated every nstlist steps rgbradii = 2 ; Dielectric coefficient of the implicit solvent gb_epsilon_solvent = 80 ; Salt concentration in M for Generalized Born models gb_saltconc = 0 ; Scaling factors used in the OBC GB model. Default values are OBC(II) gb_obc_alpha = 1 gb_obc_beta = 0.8 gb_obc_gamma = 4.85 ; Surface tension (kJ/mol/nm^2) for the SA (nonpolar surface) part of GBSA ; The default value (2.092) corresponds to 0.005 kcal/mol/Angstrom^2. sa_surface_tension = 2.092 ; OPTIONS FOR WEAK COUPLING ALGORITHMS ; Temperature coupling tcoupl = v-rescale ; Groups to couple separately tc-grps = protein ; Time constant (ps) and reference temperature (K) tau_t = 2 ref_t = 300 ; Pressure coupling Pcoupl = no Pcoupltype = Isotropic ; Time constant (ps), compressibility (1/bar) and reference P (bar) tau-p = 1 compressibility = ref-p = ; Scaling of reference coordinates, No, All or COM refcoord_scaling = No ; Random seed for Andersen thermostat andersen_seed = 815131 ; OPTIONS FOR QMMM calculations QMMM = no ; Groups treated Quantum Mechanically QMMM-grps = ; QM method QMmethod = ; QMMM scheme QMMMscheme = normal ; QM basisset QMbasis = ; QM charge QMcharge = ; QM multiplicity QMmult = ; Surface Hopping SH = ; CAS space options CASorbitals = CASelectrons = SAon = SAoff = SAsteps = ; Scale factor for MM charges MMChargeScaleFactor = 1 ; Optimization of QM subsystem bOPT = bTS = ; SIMULATED ANNEALING ; Type of annealing for each temperature group (no/single/periodic) annealing = ; Number of time points to use for specifying annealing in each group annealing_npoints = ; List of times at the annealing points for each group annealing_time = ; Temp. at each annealing point, for each group. annealing_temp = ; GENERATE VELOCITIES FOR STARTUP RUN gen_vel = yes gen_temp = 300 gen_seed = 173528 ; OPTIONS FOR BONDS constraints = all-bonds ; Type of constraint algorithm constraint-algorithm = Lincs ; Do not constrain the start configuration continuation = no ; Use successive overrelaxation to reduce the number of shake iterations Shake-SOR = no ; Relative tolerance of shake shake-tol = 0.0001 ; Highest order in the expansion of the constraint coupling matrix lincs-order = 4 ; Number of iterations in the final step of LINCS. 1 is fine for ; normal simulations, but use 2 to conserve energy in NVE runs. ; For energy minimization with constraints it should be 4 to 8. lincs-iter = 1 ; Lincs will write a warning to the stderr if in one step a bond ; rotates over more degrees than lincs-warnangle = 30 ; Convert harmonic bonds to morse potentials morse = no ; ENERGY GROUP EXCLUSIONS ; Pairs of energy groups for which all non-bonded interactions are excluded energygrp_excl = ; WALLS ; Number of walls, type, atom types, densities and box-z scale factor for Ewald nwall = 0 wall_type = 9-3 wall_r_linpot = -1 wall_atomtype = wall_density = wall_ewald_zfac = 3 ; COM PULLING ; Pull type: no, umbrella, constraint or constant_force pull = no ; NMR refinement stuff ; Distance restraints type: No, Simple or Ensemble disre = No ; Force weighting of pairs in one distance restraint: Conservative or Equal disre-weighting = Conservative ; Use sqrt of the time averaged times the instantaneous violation disre-mixed = no disre-fc = 1000 disre-tau = 0 ; Output frequency for pair distances to energy file nstdisreout = 100 ; Orientation restraints: No or Yes orire = no ; Orientation restraints force constant and tau for time averaging orire-fc = 0 orire-tau = 0 orire-fitgrp = ; Output frequency for trace(SD) and S to energy file nstorireout = 100 ; Dihedral angle restraints: No or Yes dihre = yes dihre_fc = 10 ; Free energy control stuff free-energy = no init-lambda = 0 delta-lambda = 0 sc-alpha = 0 sc-power = 0 sc-sigma = 0.3 couple-moltype = couple-lambda0 = vdw-q couple-lambda1 = vdw-q couple-intramol = no ; Non-equilibrium MD stuff acc-grps = accelerate = freezegrps = freezedim = cos-acceleration = 0 deform = ; Electric fields ; Format is number of terms (int) and for all terms an amplitude (real) ; and a phase angle (real) E-x = E-xt = E-y = E-yt = E-z = E-zt = ; User defined thingies user1-grps = user2-grps = userint1 = 0 userint2 = 0 userint3 = 0 userint4 = 0 userreal1 = 0 userreal2 = 0 userreal3 = 0 userreal4 = 0
top file (here with restraint - without is identical except it lacks the [ dihedral restraint ] section):
;
; File 'gly3.top' was generated
; By user: ms872 (1005)
; On host: nursie
; At date: Tue Oct 19 23:07:48 2010
;
; This is your topology file
; "Fresh Air, Green Hair" (Frank Black)
;
; Include forcefield parameters
#include "ffoplsaa.itp"
[ moleculetype ]
; Name nrexcl
Protein 3
[ atoms ]
; nr type resnr residue atom cgnr charge mass typeB chargeB massB
1 opls_900 1 GLY N 1 -0.9 14.0067 ; qtot -0.9
2 opls_909 1 GLY H1 1 0.36 1.008 ; qtot -0.54
3 opls_909 1 GLY H2 1 0.36 1.008 ; qtot -0.18
4 opls_906B 1 GLY CA 1 0.06 12.011 ; qtot -0.12
5 opls_140 1 GLY HA1 1 0.06 1.008 ; qtot -0.06
6 opls_140 1 GLY HA2 1 0.06 1.008 ; qtot 0
7 opls_235 1 GLY C 2 0.5 12.011 ; qtot 0.5
8 opls_236 1 GLY O 2 -0.5 15.9994 ; qtot 0
9 opls_238 2 GLY N 3 -0.5 14.0067 ; qtot -0.5
10 opls_241 2 GLY H 3 0.3 1.008 ; qtot -0.2
11 opls_223B 2 GLY CA 3 0.08 12.011 ; qtot -0.12
12 opls_140 2 GLY HA1 3 0.06 1.008 ; qtot -0.06
13 opls_140 2 GLY HA2 3 0.06 1.008 ; qtot 0
14 opls_235 2 GLY C 4 0.5 12.011 ; qtot 0.5
15 opls_236 2 GLY O 4 -0.5 15.9994 ; qtot 0
16 opls_238 3 GLY N 5 -0.5 14.0067 ; qtot -0.5
17 opls_241 3 GLY H 5 0.3 1.008 ; qtot -0.2
18 opls_223B 3 GLY CA 5 0.08 12.011 ; qtot -0.12
19 opls_140 3 GLY HA1 5 0.06 1.008 ; qtot -0.06
20 opls_140 3 GLY HA2 5 0.06 1.008 ; qtot 0
21 opls_267 3 GLY C 6 0.52 12.011 ; qtot 0.52
22 opls_269 3 GLY OT 6 -0.44 15.9994 ; qtot 0.08
23 opls_268 3 GLY O 6 -0.53 15.9994 ; qtot -0.45
24 opls_270 3 GLY HO 6 0.45 1.008 ; qtot 0
[ dihedral_restraints ]
; ai aj ak al type label phi dphi kfac power
; phi C'(n-1) - N - CA - C'
7 9 11 14 1 1 -90 75 2 1
14 16 18 21 1 1 -90 75 2 1
[ bonds ]
; ai aj funct c0 c1 c2 c3
1 2 1
1 3 1
1 4 1
4 5 1
4 6 1
4 7 1
7 8 1
7 9 1
9 10 1
9 11 1
11 12 1
11 13 1
11 14 1
14 15 1
14 16 1
16 17 1
16 18 1
18 19 1
18 20 1
18 21 1
21 22 1
21 23 1
23 24 1
[ pairs ]
; ai aj funct c0 c1 c2 c3
1 8 1
1 9 1
2 5 1
2 6 1
2 7 1
3 5 1
3 6 1
3 7 1
4 10 1
4 11 1
5 8 1
5 9 1
6 8 1
6 9 1
7 12 1
7 13 1
7 14 1
8 10 1
8 11 1
9 15 1
9 16 1
10 12 1
10 13 1
10 14 1
11 17 1
11 18 1
12 15 1
12 16 1
13 15 1
13 16 1
14 19 1
14 20 1
14 21 1
15 17 1
15 18 1
16 22 1
16 23 1
17 19 1
17 20 1
17 21 1
18 24 1
19 22 1
19 23 1
20 22 1
20 23 1
22 24 1
[ angles ]
; ai aj ak funct c0 c1 c2 c3
2 1 3 1
2 1 4 1
3 1 4 1
1 4 5 1
1 4 6 1
1 4 7 1
5 4 6 1
5 4 7 1
6 4 7 1
4 7 8 1
4 7 9 1
8 7 9 1
7 9 10 1
7 9 11 1
10 9 11 1
9 11 12 1
9 11 13 1
9 11 14 1
12 11 13 1
12 11 14 1
13 11 14 1
11 14 15 1
11 14 16 1
15 14 16 1
14 16 17 1
14 16 18 1
17 16 18 1
16 18 19 1
16 18 20 1
16 18 21 1
19 18 20 1
19 18 21 1
20 18 21 1
18 21 22 1
18 21 23 1
22 21 23 1
21 23 24 1
[ dihedrals ]
; ai aj ak al funct c0 c1 c2 c3 c4 c5
2 1 4 5 3
2 1 4 6 3
2 1 4 7 3
3 1 4 5 3
3 1 4 6 3
3 1 4 7 3
1 4 7 8 3
1 4 7 9 3
5 4 7 8 3
5 4 7 9 3
6 4 7 8 3
6 4 7 9 3
4 7 9 10 3
4 7 9 11 3
8 7 9 10 3
8 7 9 11 3
7 9 11 12 3
7 9 11 13 3
7 9 11 14 3
10 9 11 12 3
10 9 11 13 3
10 9 11 14 3
9 11 14 15 3
9 11 14 16 3
12 11 14 15 3
12 11 14 16 3
13 11 14 15 3
13 11 14 16 3
11 14 16 17 3
11 14 16 18 3
15 14 16 17 3
15 14 16 18 3
14 16 18 19 3
14 16 18 20 3
14 16 18 21 3
17 16 18 19 3
17 16 18 20 3
17 16 18 21 3
16 18 21 22 3
16 18 21 23 3
19 18 21 22 3
19 18 21 23 3
20 18 21 22 3
20 18 21 23 3
18 21 23 24 3
22 21 23 24 3
[ dihedrals ]
; ai aj ak al funct c0 c1 c2 c3
4 9 7 8 1 improper_O_C_X_Y
7 11 9 10 1 improper_Z_N_X_Y
11 16 14 15 1 improper_O_C_X_Y
14 18 16 17 1 improper_Z_N_X_Y
18 22 21 23 1 improper_O_C_X_Y
; Include Position restraint file
#ifdef POSRES
#include "posre.itp"
#endif
; Include water topology
#include "spc.itp"
#ifdef POSRES_WATER
; Position restraint for each water oxygen
[ position_restraints ]
; i funct fcx fcy fcz
1 1 1000 1000 1000
#endif
; Include generic topology for ions
#include "ions.itp"
[ system ]
; Name
Protein
[ molecules ]
; Compound #mols
Protein 1