Difference between revisions of "HMM and alignment"
Devicerandom (Talk | contribs) |
Devicerandom (Talk | contribs) |
||
| Line 4: | Line 4: | ||
* In building a profile HMM, an existing multiple alignment is given as input. | * In building a profile HMM, an existing multiple alignment is given as input. | ||
| + | * it pays to build HMMs on pre-aligned data whenever possible. | ||
| + | * Especially for complicated HMMs, the parameter space may be complex, with many spurious local optima that can trap a training algorithm. | ||
* Profile HMMs are similar to simple sequence profiles, but in addition to the amino acid frequencies in the columns of a multiple sequence alignment they contain the position-specific probabilities for inserts and deletions along the alignment | * Profile HMMs are similar to simple sequence profiles, but in addition to the amino acid frequencies in the columns of a multiple sequence alignment they contain the position-specific probabilities for inserts and deletions along the alignment | ||
Revision as of 18:42, 11 February 2013
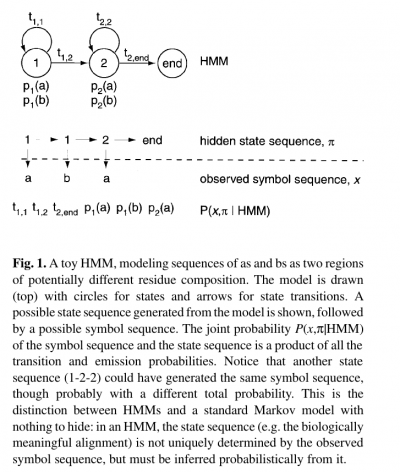
- The name ‘hidden Markov model’ comes from the fact that the state sequence is a first-order Markov chain, but only the symbol sequence is directly observed.
- Alternatively, an HMM can be built from prealigned (pre-labeled) sequences (i.e. where the state paths are assumed to be known).
- In the latter case, the parameter estimation problem is simply a matter of converting observed counts of symbol emissions and state transitions into probabilities.
- In building a profile HMM, an existing multiple alignment is given as input.
- it pays to build HMMs on pre-aligned data whenever possible.
- Especially for complicated HMMs, the parameter space may be complex, with many spurious local optima that can trap a training algorithm.
- Profile HMMs are similar to simple sequence profiles, but in addition to the amino acid frequencies in the columns of a multiple sequence alignment they contain the position-specific probabilities for inserts and deletions along the alignment
- The logarithms of these probabilities are in fact equivalent to position-specific gap penalties (Durbin et al., 1998).
- The states of the HMM are often associated with meaningful biological labels, such as ‘structural position 42’. In our toy HMM, for instance, states 1 and 2 correspond to a biological notion of two sequence regions with differing residue composition.
- Inferring the alignment of the observed protein or DNA sequence to the hidden state sequence is like labeling the sequence with relevant biological information.
- The alignment algorithm maximizes a weighted form of coemission probability, the probability that the two HMMs will emit the same sequence of residues.
- Amino acids are weighted according to their abundance, rare coemitted amino acids contributing more to the alignment score.
- Secondary structure can be included in the HMM-HMM comparison.
- We score pairs of aligned secondary structure states in a way analogous to the classical amino acids substitution matrices.
- We use ten different substitution matrices that we derived from a statistical analysis of the structure database, one for each confidence value given by PSIPRED.