Difference between revisions of "HMM and alignment"
Devicerandom (Talk | contribs) |
Devicerandom (Talk | contribs) (→How to use HHpred etc. for structure prediction) |
||
| Line 51: | Line 51: | ||
* '''1. Build a multiple sequence alignment for the target sequence''' | * '''1. Build a multiple sequence alignment for the target sequence''' | ||
| − | ** HHpred2 runs the buildali.pl script from the HHsearch 1.5.0 software package. | + | ** HHpred2 runs the ''buildali.pl'' script from the HHsearch 1.5.0 software package. |
| − | ** This script performs up to eight iterative PSI-BLAST searches | + | ** This script performs up to eight iterative PSI-BLAST searches (''note: using HHblits?'') |
| + | ** Since the most common source for corrupted PSI-BLAST alignments is the inclusion of nonhomologous segments at the ends of local sequence matches, ''buildali.pl'' prunes the ends of each sequence separately if the similarity with the profile extracted after the first search iteration falls below 1/6 bit per column. | ||
[[Category:Bioinfo notes]] | [[Category:Bioinfo notes]] | ||
[[Category:Hidden Markov models]] | [[Category:Hidden Markov models]] | ||
Revision as of 14:27, 12 February 2013
This is a collage of stuff from papers and HHpred website
Contents
HMM and alignments
Building a profile HMM
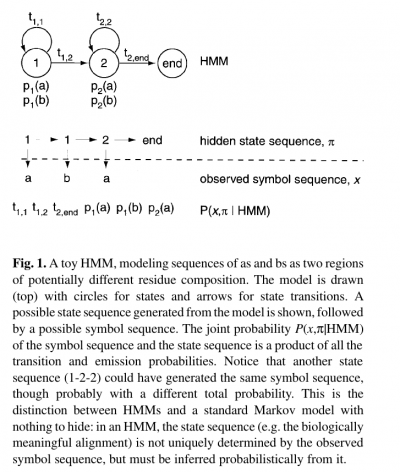
- The name ‘hidden Markov model’ comes from the fact that the state sequence is a first-order Markov chain, but only the symbol sequence is directly observed.
- Alternatively, an HMM can be built from prealigned (pre-labeled) sequences (i.e. where the state paths are assumed to be known).
- In the latter case, the parameter estimation problem is simply a matter of converting observed counts of symbol emissions and state transitions into probabilities.
- In building a profile HMM, an existing multiple alignment is given as input.
- it pays to build HMMs on pre-aligned data whenever possible.
- Especially for complicated HMMs, the parameter space may be complex, with many spurious local optima that can trap a training algorithm.
How it works inside
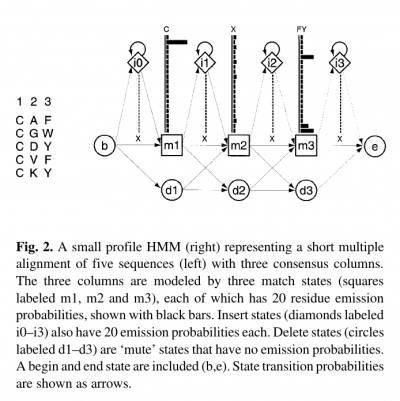
- Profile HMMs are similar to simple sequence profiles, but in addition to the amino acid frequencies in the columns of a multiple sequence alignment they contain the position-specific probabilities for inserts and deletions along the alignment
- For each consensus column of the multiple alignment, a ‘match’ state models the distribution of residues allowed in the column.
- An ‘insert’ state and ‘delete’ state at each column allow for insertion of one or more residues between that column and the next, or for deleting the consensus residue.
- Profile HMMs are strongly linear, left right models, unlike the general HMM case.
- The probability parameters in a profile HMM are usually converted to additive log-odds scores before aligning and scoring a query sequence (Barrett et al., 1997).
- The scores for aligning a residue to a profile match state are therefore comparable to the derivation of BLAST or FASTA scores: if the probability of the match state emitting residue x is px , and the expected background frequency of residue x in the sequence database is fx , the score for residue x at this match state is log(px/fx).
- The logarithms of these probabilities are in fact equivalent to position-specific gap penalties (Durbin et al., 1998).
- In contrast to parameter estimation, a suitable HMM architecture (the number of states, and how they are connected by state transitions) must usually be designed by hand.
- A maximum likelihood architecture construction algorithm exists for the special case of building profile HMMs from multiple alignments (Durbin et al., 1998).
- Insertions tend to be seen most often in surface loops of protein structures, and so have a bias towards hydrophilic residues.
- Profile HMMs can capture this information in the insert state emission distributions.
Meaning of HMM states
- The states of the HMM are often associated with meaningful biological labels, such as ‘structural position 42’. In our toy HMM, for instance, states 1 and 2 correspond to a biological notion of two sequence regions with differing residue composition.
- Inferring the alignment of the observed protein or DNA sequence to the hidden state sequence is like labeling the sequence with relevant biological information.
Alignment of HMMs
- The alignment algorithm maximizes a weighted form of coemission probability, the probability that the two HMMs will emit the same sequence of residues.
- Amino acids are weighted according to their abundance, rare coemitted amino acids contributing more to the alignment score.
- Secondary structure can be included in the HMM-HMM comparison.
- We score pairs of aligned secondary structure states in a way analogous to the classical amino acids substitution matrices.
- We use ten different substitution matrices that we derived from a statistical analysis of the structure database, one for each confidence value given by PSIPRED.
How to use HHpred etc. for structure prediction
-- From "Fast and accurate automatic structure prediction with HHpred", Andrea Hildebrand, Michael Remmert, Andreas Biegert, and Johannes Soding , Proteins 2009 --
- 1. Build a multiple sequence alignment for the target sequence
- HHpred2 runs the buildali.pl script from the HHsearch 1.5.0 software package.
- This script performs up to eight iterative PSI-BLAST searches (note: using HHblits?)
- Since the most common source for corrupted PSI-BLAST alignments is the inclusion of nonhomologous segments at the ends of local sequence matches, buildali.pl prunes the ends of each sequence separately if the similarity with the profile extracted after the first search iteration falls below 1/6 bit per column.